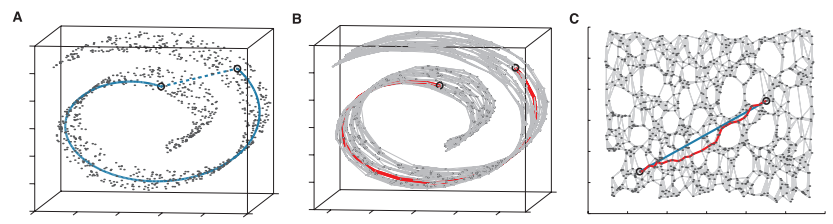
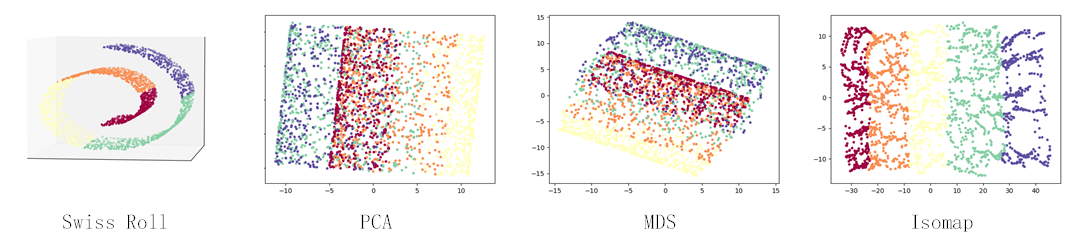
from IsomapCuda import *
from DataUtils import *
import getopt,sys
GPU_MEM_SIZE = 512
def Isomap(dataSet,outfile,srcDims,trgDims,k,eps=1000000000., CIsomap=False):
"""
Classical isomap
"""
knnRefs,knnDists,knnm = KNN(dataSet,k,eps,srcDims)
mdists = []
if CIsomap:
mdists = C_Isomap(knnDists,knnm,k)
pathMatrix = APSP(knnRefs,knnDists,knnm,eps)
del knnRefs
del knnDists
del knnm
normMatrix = NormMatrix(pathMatrix,mdists)
del pathMatrix
del mdists
embedding = QEig(normMatrix,trgDims)
del normMatrix
return embedding
def NMIsomap(dataSet,outfile,srcDims,trgDims,k,eps=1000000000., saveSteps = False):
"""
Non-Metric Isomap
"""
knnRefs,knnDists = loadSplitTable(KNN(dataSet,k,eps,srcDims))
pathMatrix = APSP(knnRefs,knnDists,eps)
del knnRefs
del knnDists
if saveSteps:
saveTable(pathMatrix,outfile[:-4]+'_distances.csv')
origDims = len(pathMatrix)
rankMatrix = RankMatrix(pathMatrix)
del pathMatrix
embedding = NMDS(rankMatrix, loadMatrix(dataSet)[:,:trgDims], origDims, trgDims)
return embedding
if __name__ == '__main__':
arg_values = ['nonmetric=','outdims=','indims=','if=','of=','k=','eps=','help','h']
optlist, args = getopt.getopt(sys.argv[1:], 'x', arg_values)
trgDims = 3
srcDims = 10000000000
k =6
eps = 1000000000.
infile='swissroll.csv'
outfile='embedding.csv'
nonmetric=False
for o in optlist:
if o[0].strip('-') == 'outdims':
trgDims = int(o[1])
for o in optlist:
if o[0].strip('-') == 'indims':
srcDims = int(o[1])
for o in optlist:
if o[0].strip('-') == 'if':
infile = o[1]
for o in optlist:
if o[0].strip('-') == 'of':
outfile = o[1]
for o in optlist:
if o[0].strip('-') == 'k':
k = int(o[1])
for o in optlist:
if o[0].strip('-') == 'nonmetric':
if o[1].strip(' \r\n\t') == 'True' or o[1].strip(' \r\n\t') == 'true':
nonmetric = True
for o in optlist:
if o[0].strip('-') == 'help' or o[1].strip('-') == 'h':
print "The following commands are available:"
print "\t--if=inputfile\tDefaults to swissroll.csv"
print "\t--of=outputfile\tDefaults to embedding.csv"
print "\t--k=k_nearest_neighbours\tDefaults to 12"
print "\t--outdims=embedding_dimensions\tDefaults to 3"
print "\t--indims=input_dimensions\tDefaults to all in the input file"
print "\t--nonmetric\tEnables non-metric MDS embeddings"
result = None
if not nonmetric:
result = Isomap(infile,outfile,srcDims,trgDims,k,eps,False)
else:
result = NMIsomap(infile,outfile,srcDims,trgDims,k,eps,False)
saveTable(result,outfile)
|